We’re meeting again at Timelab this Wednesday July 19th at 8pm. We’ll go over some practical stuff for starting the lab work in August. As posted before, we are going to the lab at UGent where we can do work with the plasmids on August 2nd, 7pm.
@flinty will be there to share with us her limitless knowledge on making lab magic happen 
Maybe a visit to the Gentse Feesten afterwards? Let’s see!
Notes: basic plasmid mechanics
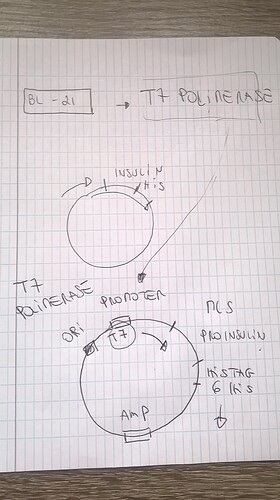
@flinty and I went over the vector, how it works and the implications for the protocols. I wanted to share this for those who are fully clear on the mechanics yet. Below is a simple schematic and here’s a more detailed vector map.
Basically it comes down to this: for expressing proinsulin, the bacteria needs T7 polymerase (which is a molecule needed for turning DNA into a protein) and a spot to bind to the plasmid DNA. The bacteria has to produce T7 polymerase itself (that’s why we need a specific type of bacteria, such as BL21 or TG1, that does this). We control production through making the binding spot available or not.
The T7 polymerase can only bind to the T7 promotor, which leads to production of proinsulin, when the binding spot is not repressed. Adding IPTG, which works on the “lac” part on the plasmid, makes sure the T7 promotor is not repressed. IPTG is our induction agent in this case: when we add it, proinsulin production starts.
The “his tag” is for purification of the protein after it was expressed. The amp gene is for ampiciline resistance so that when we add ampiciline (an antibiotic) to the growth medium, we are sure the surviving bacteria have our plasmid (those bacteria that don’t have the plasmid are not resistant).
Finally, we went over the required materials one last time and we’re good to go for starting experiments in August when we have access to the lab!